Identifying Fungi Spores, Yeast, Bacteria by Opto-Electronic Imaging and Image Processing and Identification for Protecting Human Health
CurrentTrends in Biomedical Engineering & Biosciences Juniper Publishers
Authored by Petra Perner*
Abstract
Human beings are exposed every day to bio-aerosols in their personal and/or professional life. The European Commission has issued regulations for protecting employees in the workplace from biological hazards. Airborne fungi can be detected and identified by an image-acquisition and interpretation system. In this paper we present recent results on the development of an automated image acquisition, sample handling and image-interpretation system for airborne fungi identification. We explain the application domain and describe the development issues. The development strategy and the architecture of the system are described and results are presented.
Keywords: Health monitoring; Microscopic image acquisition; microbiological sample handling; image analysis; image interpretation; case-based object recognition; case-based reasoning
Introduction
Airborne microorganisms are ubiquitously present in various indoor and outdoor environments. The potential implication of fungal contaminants in bio-aerosols on occupational health has been recognized as a problem in several working environments. The exposure of workers to bio-aerosols is a concern especially in composting facilities, in agriculture, and in municipal waste treatment. The European Commission has therefore issued guidelines protecting employees in the workplace from airborne biological hazards. In fact, the number of incidents of building-related sickness, especially in offices and residential buildings, is increasing. Some of these problems are attributed to biological agents, especially to airborne fungal spores. However, the knowledge of health effects of indoor fungal contaminants is still limited. One of the reasons for this limitation is that appropriate methods for rapid and long-time monitoring of airborne microorganisms are not available. In addition to the detection of parameters relevant to occupational and public health, in many controlled environments the number of airborne microorganisms has to be kept below the permissible or recommended values, e.g. in clean rooms, in operating theaters, and in domains of the food and pharmaceutical industry. Consequently, the continuous monitoring of airborne biological agents is a necessity for the detection of risks to human health as well as for the flawless operation of technological processes.
Airborne microorganisms are ubiquitously present in various indoor and outdoor environments. The potential implication of fungal contaminants in bio-aerosols on occupational health has been recognized as a problem in several working environments. The exposure of workers to bio-aerosols is a concern especially in composting facilities, in agriculture, and in municipal waste treatment. The European Commission has therefore issued guidelines protecting employees in the workplace from airborne biological hazards. In fact, the number of incidents of building-related sickness, especially in offices and residential buildings, is increasing. Some of these problems are attributed to biological agents, especially to airborne fungal spores. However, the knowledge of health effects of indoor fungal contaminants is still limited. One of the reasons for this limitation is that appropriate methods for rapid and long-time monitoring of airborne microorganisms are not available. In addition to the detection of parameters relevant to occupational and public health, in many controlled environments the number of airborne microorganisms has to be kept below the permissible or recommended values, e.g. in clean rooms, in operating theaters, and in domains of the food and pharmaceutical industry. Consequently, the continuous monitoring of airborne biological agents is a necessity for the detection of risks to human health as well as for the flawless operation of technological processes.
In the system described here, contaminated air containing bio-aerosols is collected in a defined volume via a carrier agent. Bio-aerosols are recorded by an image-acquisition unit, counted, and classified. Their nature is determined by means of an automated image-analysis and interpretation system. Air samples are automatically acquired, prepared and transferred by a multi-axis servo-system to an image-acquisition unit comprised of a standard optical microscope with a digital color camera. This part of the system is described in Section 2. To obtain a sufficient image quality, special requirements have to be fulfilled by the image- acquisition unit which will be described in Section 3.
The variability of the biological objects is very broad. Given the constraints of the image acquisition, this variability is found in the appearance of the objects as well. There are no general How to cite this article: Petra P. Identifying Fungi Spores, Yeast, Bacteria by Opto-Electronic Imaging and Image Processing and Identification for 002 Protecting Human Health. Curr Trends Biomedical Eng & Biosci. 2018; 11(2): 555806. DOI: 10.19080/CTBEB.2018.11.555806. features allowing one to discern the type of the detected fungi. In the system employed here, images are stored, and a more generalized description for the different appearances of the same objects is used. We will describe this novel case-based reasoning approach for the image analysis and its interpretation in Section 4. Finally, we will summarize our work in Section 5.
General comments on the image analysis of microorganism
Classification of airborne fungal spores from environmental samples presents the image analyst with inherent difficulties. Most of these difficulties concern the automatic identification of microorganism in general [2]. For example, the types and numbers of objects (different fungal species) that may be present in any one air sample are both unknown and effectively unlimited. Also, intra-species variation of characteristics (such as size, color or texture of spores) can be large and may depend on several factor. Furthermore, the bulk size of two targeted species may be an order of magnitude or more apart, making it difficult to decide e.g. on an optical magnification setting. The dynamic and variable nature of the microorganism thus presents a formidable challenge in regard to the design of a robust image interpretation system with the ideal characteristics of high analysis accuracy but wide generalization ability. The difficulties can be summarized as follow:
Intra-species variation due to natural phenomenon,i.e., life-cycle, environmental effects
The dynamic nature of living organisms results in properties such as size or color of the microorganism being statistically variable. Different growth condition of microorganism may result in uncharacteristically large or small specimens – resulting in data outliers. Ultimately, under these circumstances the classification accuracy of an image interpretation system will rely on the training database capturing as much of this variability as possible.
Intra-species variation due to predation, fragmentation etc.
Often atypical characteristics occur due to predation, environmental factors, or aging.
To stain or not to stain?
Many species appear clear/opaque at the resolutions used, making imaging and analysis very difficult. Staining can help to increase the resolution of the fungal material and to distinguish between viable and non-viable organisms. Depending on the application different stains have to be used. At present 10-20 different stains are frequently used for staining fungal spores. They include “all-purpose”-stains such as lactophenol cotton blue which stains fungal elements blue. The staining procedure takes only 1 to 2 minutes. The application of fluorescence stains allows to discriminate between living and dead cells. However the use of epifluorescence microscopy in an automated system is more expensive and requires additional hardware. While it is common to stain specimen samples prior to analysis, staining puts special demands on an automated sample handling, image acquisition system and image interpretation system.
Choosing an appropriate optical resolution for imaging specimens
The wide variation of the size of targeted species necessitates a choice of optical magnification that may not be optimal for any species. For example, to analyse the fine internal structures of species such as Wallemia sebi, a 1000x magnification would be required. Fusarium spores are the largest spores among the spores considered in this study. They would require only a 200x magnification instead of a 1000x magnification.
Imaging 3-dimensional objects
The spore is a 3-dimensional object. Imagine a spore which has an ellipsoid shape. Depending on its position, the object can appear as a round object or as an elongated object in a 2-D image. Many species have a significant length in the third dimension - often greater than the depth-of-field of the imaging device-making their representation as a 2-D image difficult. As such, significant areas of the specimen will be out of focus. If only one kind of specimen appears in an image focusing may not be so difficult. However, in a real air sample different specimen can appear. In this case, a single focus level may not be sufficient. Different levels of focus may be necessary which will result in more than one digital image for one sample.
How to get a clean sample from the air sample?
Samples of bioaerosols will contain a wide range of objects (organic and inorganic particles). Filters will be needed to remove particles larger than the objects of interest. But this will generally not prevent the image from containing non-targeted species. Non- targeted species/objects will generally need to be classified. Normally the sample should be covered by water and a cover glass. To realize this in an automated handling system is not easy since handling glass by means of handling devices is difficult.
Related Works
Several case studies have been done on identifying fungi or other microorganism. In [3], an image analysis method was described for the identification of colonies of nine different Penicillium species as seen after growth on a standard medium. In [4], a study of image analysis based on fluorescence microscopy images was described for the improvement of the exposure assessment of airborne microorganism. Semiautomatic image analysis techniques were applied to segment the contour of fungal hyphae in [5]. Yeast cells were analyzed by image analysis techniques in [6]. Different Fusarium species macroconidia were analyzed in [7]. The work aimed at designing an automated procedure for collecting and documenting microscopic pictures of Fusarium conidia, determining various morphological parameters and statistically evaluating the effectiveness of those characteristics in differentiating the most important pathogenic Fusarium species occurring on wheat in Germany
The work which is most closely related to our work is that described in [8]. The ability of an image analysis routine to differentiate between spores of eleven allergenic fungal genera was tested using image analysis based on seven basic and up to 17 more complex features, extracted from digitized images. Fungal spores of Alternaria, Cladosporium, Fusarium, Aspergillus, Botrytis, Penicillium, Epicoccum, Exserohilum, Ustilago, Coprinus and Psilocybe were examined in a series of experiments designed to differentiate between spores at the genus and species level. No specific algorithm for image enhancement and image segmentation is described in this work. It appears that only the feature measurement has been automated. The object area was labelled interactively. From the fungal spores seven basic features including length, width, width/length ratio, area, form factor (circularity), perimeter and roundness, and 17 more complex features including equivalent circular diameter, compactness, box area, radius, modification ratio, sphericity, convex hull area, convex hull perimeter, solidity, concavity, convexity, fibre length, fibre width were extracted. Linear and quadratic discriminant analyses were used for classification. It is interesting to note that the authors created a sufficiently large database of fungi spores for their analysis. The number of spores used for this study ranges from 200 to 1000 samples. The classification accuracy according to a particular class ranged from 56% to 93% for genera comparison and from 26% to 97% for species comparison. The results showed that not for all classes the right features for classification were selected. Rather, it appeared that all common features that are known in pattern recognition for the description of a 2-D objects were applied to the images. No specific features have been developed that describe the properties of the different fungi genera and species. For example, considering specie Fusarium, the septation is a highly discriminating features but no such description was included in the feature list [8].
A number of successful case studies have been conducted to automate the identification of fungi and microorganism in general. In these studies, imaging methods for microorganism, automatic focusing methods, image analysis, feature description and classification have been developed. Most of these studies used 500x to 1,500x magnification for image acquisition. The most used feature descriptors are the area size and the shape factor of circularity. The color information was used only in [3], and was neglected in all other studies. Not all publications included microscopic images of the microorganism; therefore, we cannot evaluate the quality of the images. In most of the cases, the digitized images were not highly structured. The objects and the background appeared more or less homogenous allowing to apply a simple thresholding technique for image segmentation. In general, these studies are characterized by applying standard image analysis and feature extraction procedures to the images. Neither a specific feature set for fungi identification has been developed nor a good feature set for the description of microorganism has been found yet as evidenced by [7,8].
The difference to our work is that in most of these studies images are created for only one specie and not for a variety of different species, except for the work in [8]. The creation of digitized images for a variety of different species is much harder since the species differ in size and dimension and, therefore, the selection of an optical resolution that will show the image details of the different species in sufficient resolution is not easy. Also, the image analysis is much more difficult since for all the different objects a sufficient image quality should be reached after image segmentation.
Development Issues
We decided to start the development of our system based on a data set of fungi spore images taken in the laboratory under optimal conditions and constant climate conditions. The data set should represent the prototypical appearance of the different kind of fungi strains and serve as gold standard. The objects in the images are good representatives of the different kinds of fungal spores cultured under optimal conditions and constant climate conditions. However, as it can be seen from the images of Alternaria alternata and Ulocladium botrytis none of the objects in the image looks like another. There is no clear prototypical object. We can see a high biological variability and also younger and older representatives of the fungal strains. Depending on the image acquisition conditions we see objects from the side and from the top and this influences the appearance of the objects. Generalization about the objects cannot be done manually; rather, each case that appears in practice should be stored in the system and the system should learn more generalized descriptions for the different appearance of the same objects over time. All this suggests that a case-based reasoning approach for the image interpretation [9] should be taken rather than a generalized approach. Case-Based Reasoning [10] is used when generalized knowledge is lacking. The method works on a set of cases previously processed and stored in a case base. A new case is interpreted by searching for similar cases in the case base. Among this set of similar cases the closest case with its associated result is selected and presented on a display.
For the kind of images created in the laboratory we have to develop an image analysis procedure. It is then necessary to describe the images by image features and to develop a feature extraction procedure which can automatically extract the features from the images. The features and the feature values extracted from the images together with the name of the fungal spores make up an initial description of the data. We do not know if all image features are indeed necessary. However, we extract as many image features as possible from the images that appear meaningful in some way to ensure that we can mine the right case description from this database. From this initial description of the data we need to identify good representative descriptions for the cases by using case mining methods [10]. Based on this information we will generate the case-based reasoning system. After reaching a sufficient classification accuracy we will start to include real air samples into the system by adapting the prototypical representations of fungi spores to the real ones.
System requirements
The system to be developed should allow to collect dust and biological aerosols in well- defined volumes over microscope slides, deposit them there, image them with an appropriate method and count and classify them with an automated image analysis and interpretation method, in order to determine the following parameters from the images:
a) Total number of airborne particles
b) Classification of all particles according to their size and shape
c) Classification of biological particles according to their size and shape, e.g. spores, fragments of fungal mycelia, and fragments of insects
d) Number of respirable particles
e) Total number of airborne particles of biological origin
f) Number of dead particles of biological origin
g) Number of viable and augmentable particles of biological origin
h) Identification of species or genera exploiting the characteristic shapes of spores and pollen
i) Proportion of airborne abiotic and biotic particles
j) Proportion of dead and viable airborne microorganisms.
At the beginning of the project the following requirements concerning the optical and the mechanical system were defined:
a) Color images should be produced in order to facilitate the separation of dead and living objects.
b) It should be possible to generate images in at least three defined depths of field.
c) A marker liquid like lactophenol should be used to further enhance the separation of dead and living objects (blue color for living objects). For this purpose a cover slip is necessary in order to uniformly distribute the marker drop on the object slide.
d) The object slide should be covered with an adhesive in order to fix the airborne germs.
Six fungal strains representing species with different spore types were identified as important species in different environments (Table 1) by our industrial project partner Jena Bios GmbH. A database of images from the spores of these species was produced and was the basis of our development. The number of imaged spore per species was about 30-50. Since no commercial system was known fulfilling all requirements, a corresponding system was developed which is described in what follows.
The automated imaging system
The microscopic image-acquisition system: Following the specifications given in Section 2 we developed an automated sample-handling and digital image-acquisition system for taking microbiological material from air samples. An existing optical Leitz microscope was upgraded and its hardware expanded. A lens from Olympus with a magnification of 60X and a numerical aperture of 0.7 was used. Its focal length of 1.7mm provided sufficient clearance between the lens and the object slide including the cover glass to avoid collisions due to their variability in thickness. The lens was inserted in an autofocusing device from Physik Instrumente (PI, Karlsruhe, Germany) which was mounted on the lens revolver. A motorized xy-table from Märzhäuser (Wetzlar, Germany) with a controller was used to arbitrarily shift the object slide in both x and y direction. For the digital image acquisition a 1.4M pixel color digital camera from Soft Imaging System (SIS, Münster, Germany) was used. Our estimates showed that a pixel number larger than 1.4Mpixel is sufficient for the given magnification. Figure 1 demonstrates that the optical resolution is sufficient to recognize details in spores like Ulocladium.
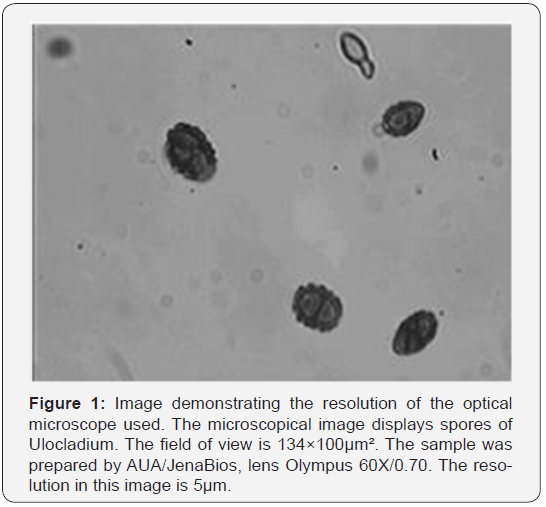
The functions of image acquisition and image storage, movement of the specimen in x and y direction, and autofocusing in z-direction are controlled by the Analysis Pro software from SIS. A pattern of images at any image position can be freely programmed and stored in a macro-code. This holds true also for the number of images to be captured. If necessary it is possible to capture automatically images at different depths of focus around the optimum position. By the automatic shading correction, the effect of an inhomogeneous illumination of the object can be removed.

The automatic sample-acquisition and handling system: The following chapter describes the main units and functions of the demonstration set-up realized in the course of the project. A stock of special object slides covered with a sticky layer and obtained from Umweltanalytik Holbach [11], (Figure 2) is kept in a slide storage. A sliding gripper takes the lowest slide in the storage and transports it into the slit impactor obtained from Umweltanalytik Holbach (Figure 3). The object slides are separated by distance holders with a corresponding recess, in order to avoid sticking between the slides. The distance holder is removed by the same gripper, now moving in opposite direction and depositing the distance holder into a box. The distance holders can be used again when the slide deposit is reloaded.

In the slit impactor (Figure 3), the air, potentially containing airborne germs, is guided onto the sticky area of the object slide by the air stream generated by an air pump. After a few tens of seconds adjustable appropriately, the pump is switched off and the object slide is transported to the pipetting unit driven by the dosing pump (Cavro XL 3000 obtained from Tecan Systems San Jose, Ca, USA). To achieve this, the object slide has to change its transporting axis and thus its direction of movement. From a thin nozzle one drop of lactophenol is deposited on the sticky area of the object slide. The object slide is afterwards transported through the coordinate origin to the cover-slip gripper unit. This gripper acts as a low- pressure sucker and takes one cover glass from the deposit and places it with one edge first on the object slide. Then the cover glass is allowed to drop down on the object slide and flattens the drop so that it will be distributed all over the sticky area forming a thin layer. In this way the airborne germs collected on the sticky layer are immersed in the lactophenol. In lactophenol living germs take on a blue color. The object slide is then transported back to the coordinate origin where it again changes its direction of movement by 90° and is transported to the xy-table of the microscope where the slide is received and directly transported into a position underneath the lens. To this end, an additional module was integrated into the AnalySIS Pro software. It controls the manual or automated shift of the xytable between the image-acquisition position under the lens and the loading position, where the object slide is shifted from the object-slide preparation unit to the xy-table. After the object slide has reached the image acquisition position, the microscope camera then takes the images at the programmed slide positions after auto-focusing of the microscope lens at each position. The cycle of shifting the xy-table to the defined positions, auto focusing, image acquisition and storage is programmable in a macro-code integrated into the Analysis Pro software. This can also be done for other procedures like shading correction or image acquisition at different z- positions. After having finished the imaging sequence, the slide is transported away from the xytable with a special arm and drops into a box. While the image grabbing procedure by the microscope unit is still under way, the object-slide preparation unit already starts with the preparation of a new object slide.

The object-slide preparation and manipulation is performed by a hardware controller and by custom software written in C++. The transfer from the Analysis Pro software to the C++ software and vice versa is controlled by a communication protocol as interface between both software units. Altogether six different mechanical axes have to be handled, not counting the axes of the xy-table (Figure 4). The unit for object-slide preparation and the expanded microscope are shown in Figure 5.
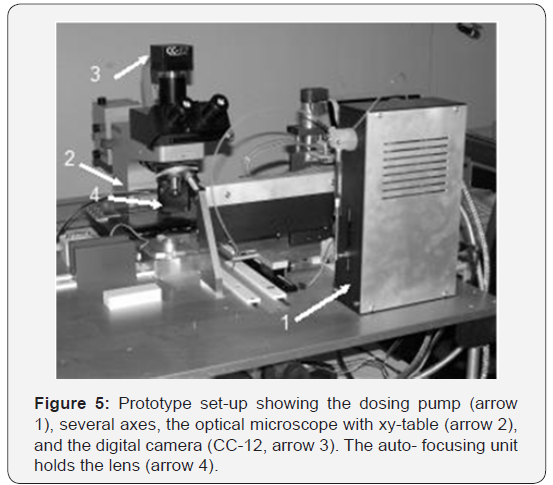
Image analysis
Once an image has been taken it is transferred to the imageanalysis unit for further processing. We will describe the overall architecture of the system [13] and its single components in the next sections.
The architecture
The architecture of the system is shown in Figure 6. Objects are recognized in the microscopic image by a case-based objectrecognition unit [14]. This unit has a case-base of shapes (case base_1) for fungi spores and determines on a similaritybased inference if there are objects in the image that have a similar shape as the ones stored in the case base. In this case the objects are labeled and transferred for further processing to the feature-extraction unit. To ensure proper performance of this unit, the general appearance of the shapes of the fungi spores must be learned. To this end we have developed a semiautomated procedure [14-20] that allows acquisition of the shape information from the raw image data and learning of groups of shape-cases and general shape-cases. A more detailed description of the case- based object-matching unit can be found in Section 4.2.
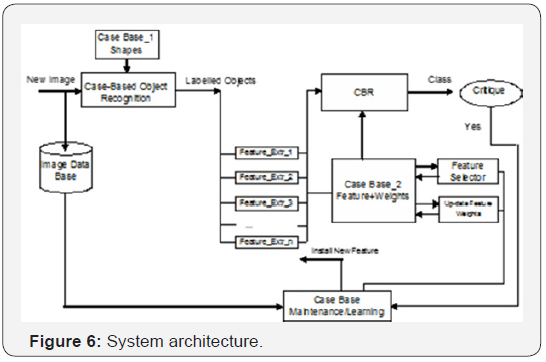
The feature-extraction procedures are based on the knowledge of an expert. Note that a particular application requires special feature descriptors. Therefore not all possible feature- extraction procedures can be implemented in such a system from the beginning. Our aim was to develop a special vocabulary and the associated feature-extraction procedures for application on fungi identification, as described in Section 4.3.
Based on the feature description, the second case-based reasoning unit determines the type of the fungi spore. This unit employs a prototype-based classifier [20]. It initially works based on prototypical cases that were selected or created by the expert. It can learn with time the different appearances of the fungi spores. The special features of this unit ensure its proper performance. It can learn the relevant prototypes from the subjectively selected set of prototypes, as well as create new prototypes. It can also learn the importance of the features of the cases. The final result of the system will be the identification of the fungi spores that appear in the image and the number of these spores. The result is shown on the display of the system and saved in a file, together with the date and time of data acquisition.
Suppose that fungi species are wrongly identified by the system. Then a case-based maintenance process will start. First the system developer must check whether new features have to be acquired for each case, or whether the whole case representation should be updated based on the learning procedures. The feature weights are learnt, as well as a subset of relevant features (see Section 4.4). To acquire new features means that necessary feature- extraction procedures have to be developed and that for all cases the new features have to be calculated and input into the existing case description. Therefore, the digital images acquired so far are retained in the image-data base. Then, the case representation as well as the index structure must be updated. This ensures that we can generate step-by-step a system that can describe the variability of the different biological objects that may appear.
Case-based object recognition
The objects in the image are highly structured. Our study has shown that the images specified in Table 1 cannot be segmented by thresholding. The objects in the image may be occluded, touching, or overlapping. It can also happen that only part of the objects appears in the image. Therefore we decided to use a case-based object recognition procedure [14] for the detection of objects in the image.
A case-based object-recognition method uses cases that generalize the original objects and compares them with the objects of the image. During this procedure a score is calculated that describes the quality of the fit between the object and the case. The case can be an object model which describes the inner appearance of the object as well as its contour. In our case the appearance of the objects as a whole can be very diverse. The shape seems to be the feature that generalizes the objects. Therefore, we decided to use contour models. We do not use the gray values of the model, but instead the object’s edges. For determining the score of the match between the contour of the object and the case, we use a similarity measure based on the scalar product that measures the average angle between the vectors of the template and the object.

1(A): from culture collection of JenaBios GmbH, Jena, Germany. 2(B): from the fungal stock collection of the Institute of Microbiology, University of Jena, Jena, Germany.
Case-base generation
The acquisition of the case is done semi-automatically. Prototypical images are shown to an expert. The expert manually traces the contour of the object by means of the cursor of the computer. Afterwards the number of contour points is reduced for data-reduction purposes by interpolating the marked contour by a first-order polynom. The marked object shapes are then aligned by the Procreates Algorithm [15]. From the sample points the direction vector is calculated. From a set of shapes, general groups of shapes are learnt by conceptual clustering which is a hierarchical incremental clustering method [16]. The prototype of each cluster is calculated by estimating the mean shape [16] of the set of shapes in the cluster and is taken as a case model.
Results for Case-Based Object Recognition
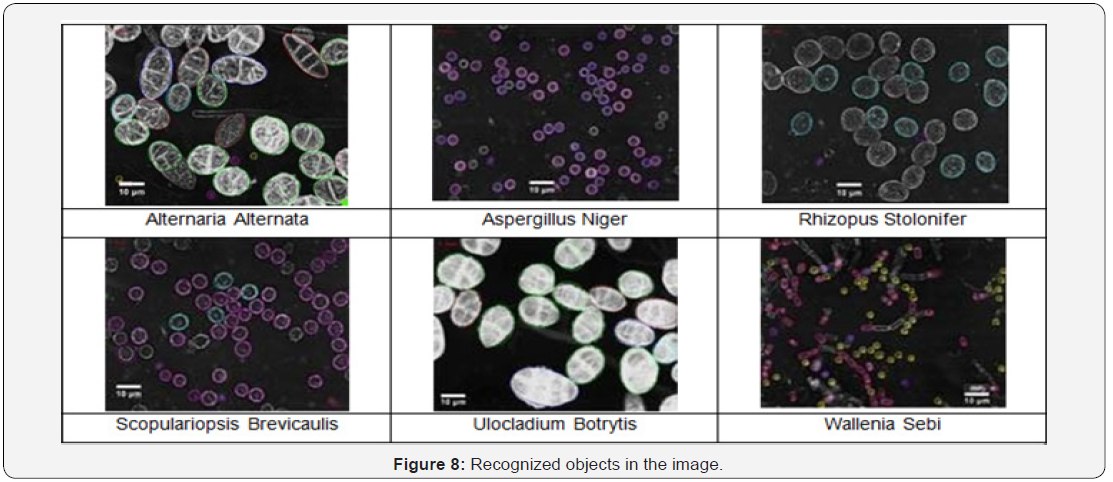
We had a total of 10 images for each class at our disposal. From this set of images two images were selected for case generation. In these two images there were approx. 60 objects. These objects were labeled and used for the case generation according to the procedure as described in Section 4.2.1. The result was a data base of cases. These cases were applied to the image for the particular class (Figure 7).
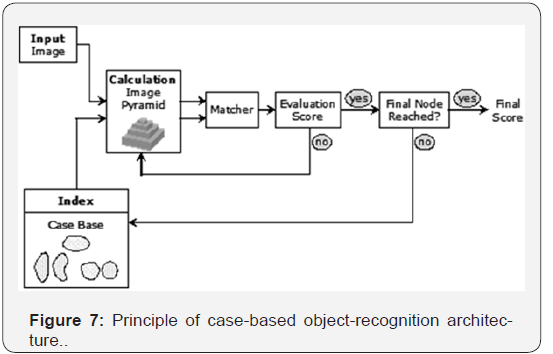
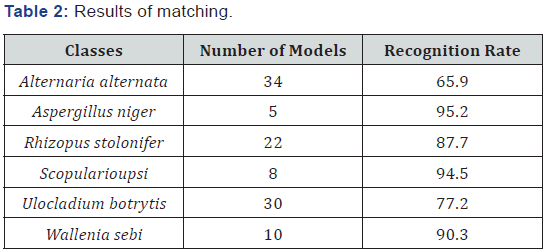
The threshold for the score was set to 0.8. We calculated the recognition rate as the number of objects that were recognized in the image to the total number of objects in the images. Note that the recognition rate can be higher than 100%, since our matching procedure also fires in image regions where no objects are present due to background noise. The aim is to configure the case-based object-recognition unit in such a way that the number of false alarms is low. The results of the matching process are shown in Figure 8 & 9. The highest recognition rate can be achieved for the objects Aspergillus niger and Scopularioupsi, since the shape of these objects does not very much. This is also expressed by the number of models, see Table 2. These classes have the lowest number of cases. For those classes where the variation of the shape of the objects is high, the number of the cases is also high. The recognition rate shows that we did not have enough cases to recognize the classes with a good recognition rate (see Ulocladium botrytis and Alternaria alternata). Therefore, we needed to increase the number of cases. For this task we developed an incremental procedure for the case acquisition in our tool. Objects that have not been recognized well will be displayed automatically for tracing and then the similarity to all other shapes will be calculated. The clustering will be done in anincremental fashion as well [16]. This procedure will ensure that we can learn the natural variation of the shape during the usage of the system.
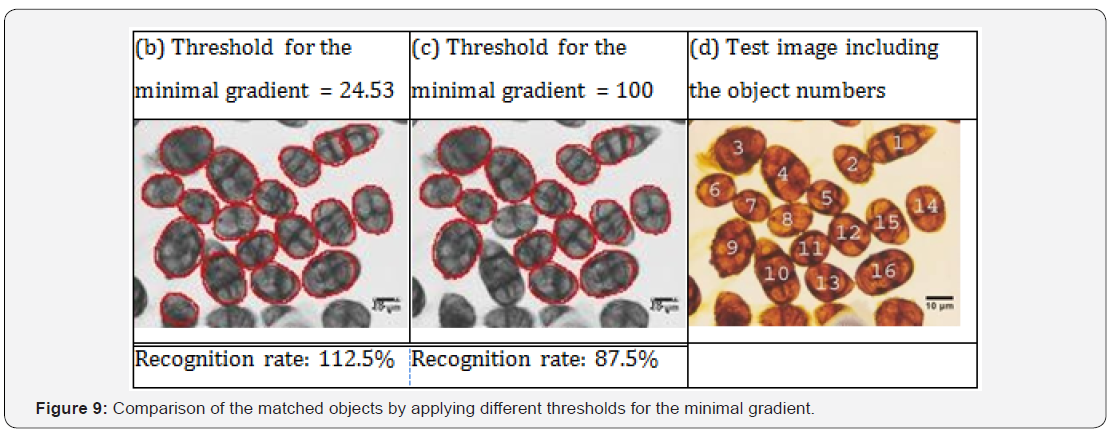
Case description and feature extraction
We choose an attribute-value pair-representation for the case description. The case consists of the solution, i.e., the type of fungi spores and the features describing the visual properties of the object (see Figure 9). From each recognized object a set of features is extracted. One feature is the case number which represents the shape of the object, the similarity score between the actual shape and the shape in the case base, the size of the object, various gray- scale features, and the texture inside the object. For the description of the texture we use our texture descriptor based on random sets described in [17].
Classification
Our case-based reasoning procedure to recognize spores relies on prototype-based classification schemes [21]. Usually such schemes are generalized from a set of single cases. Here, we have prototypical cases represented as images that were selected by humans. This means that, when building our system, we start from the top and have to collect more information about the specific class during usage of the system. Since a human has selected the prototypical images, his decision on the importance of an image might be biased; moreover selecting only one image might be difficult for a human. He can have stored more than one image as prototypical images. Therefore, we need to check the redundancy of the many prototypes for one class before taking them all into the case base. According to this consideration, our system must fulfill the following functions:
I. Classification based on the nearest neighbor rule
II. Prototype selection by a redundancy-reduction algorithm; Feature weighting to determine the importance of the features for the prototypes
III. Feature-subset to select the relevant features from the whole set of the respective domain.
The classification method is based on the nearest-neighbor rule. Since the prototypes are available at the same time, we choose a decremental redundancy-reduction algorithm proposed by Chang [18] that deletes prototypes as long as the classification accuracy does not decrease. The feature-subset selection is based on the wrapper approach [19] and an empirical feature-weighting learning method [20] is used. Furthermore, cross validation is used to estimate the classification accuracy. The prototype selection, the feature selection, and the featureweighting steps are performed during each run of the crossvalidation process. This rule classifies x in the category of its nearest neighbor [20].
More precisely, we call xn ∈ {x1, x2,..... xi,.... xn} a nearest neighbor to x if , min (xi, x) = d (ξv, ξ) where i =1,2,…n. The nearest neighbor rule classifies x into category Cn where xn is the nearest neighbor to x and xn belongs to class Cn.
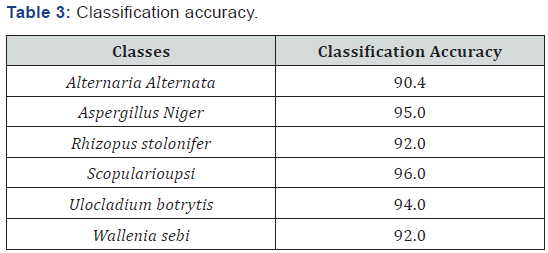
For the k-nearest neighbor we require k-samples of the same class to satisfy the decision rule. As a distance measure, we use the Euclidean distance. The recognition rate was evaluated on a data base of 50 samples for each class based on crossvalidation. The result is shown in Table 3. Based on this result, we can conclude that the classification accuracy is higher than the recognition rate for some classes. This means that it is more difficult to recognize the objects that are most likely to be fungi spores than to classify them based on the extracted features.
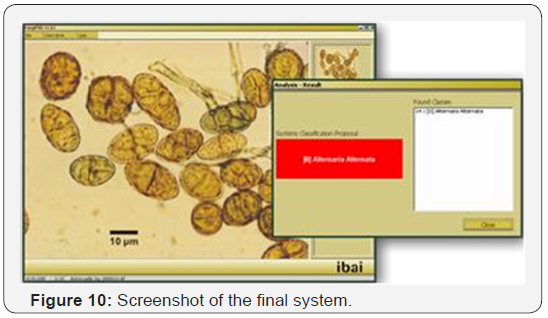
A print-out of a result obtained by the system described in this paper is shown in Figure 10. In the display the operator will find the acquired image in one window and in the other window the determined fungi spores and their total number. The system called Fungi PAD correctly identified the name of the fungi spores and their number.
Conclusion
In this paper a system for an automated image acquisition and analysis of hazardous biological material in air is described. It consists of an image-acquisition unit, its sample-handling hardware, and the image-interpretation system. The samplehandling and image-acquisition unit collects the airborne germs, deposits them on an object slide, disperses them with a marker fluid, and takes digital images of the germs in a programmable pattern. The stored images are analyzed in order to identify the germs based on a novel case-based object- recognition method. The case generation is done semi-automatically by manually tracing the contour of the object, by automated shape alignment and by shape clustering, and eventually by prototype calculation. Based on the acquired shape cases, the object-recognition unit identifies objects in the image that are likely to be fungi spores. The further examination of labeled objects is done by calculating more distinct object features, from which a prototype- based classifier determines the kind of fungi spores. After all objects have been classified by their type, the number of one type of fungi spores is calculated and displayed for the operator on the computer screen.
The recognition rate is good enough for on-line monitoring of environments. The final information can be used to determine contamination of environments with biological hazardous material. It can be used for health monitoring as well as for process control.
Acknowledgement
This project has been sponsored under the High-Tech Strategy of the Federal Republik of Germany under the grant number 16IN0147 entitled The “Development of Novel Image Analysis and Interpretation Methods for Airborne Hazardous Materials-BIOGEFA”. We appreciate the great financial support that allowed us to do and explore substantial work on that subject.
To
Know More About Current Trends in Biomedical Engineering & Biosciences Please
Click on:
https://juniperpublishers.com/ctbeb/index.php
Comments
Post a Comment